
A GPS for the brain and so much more
Scientists use NeuroInfo to help navigate the brain and compare findings across labs
Reproducibility has always been a primary goal in science. But the human effort involved in replicating a research study and analyzing the results, can be considerable. NeuroInfo® is a revolutionary new tool that scientists are using to register whole slide images into a standardized mouse brain atlas in an easy, automated way. Images and subsequent measurements can then be cross-referenced against findings from a myriad of other studies.
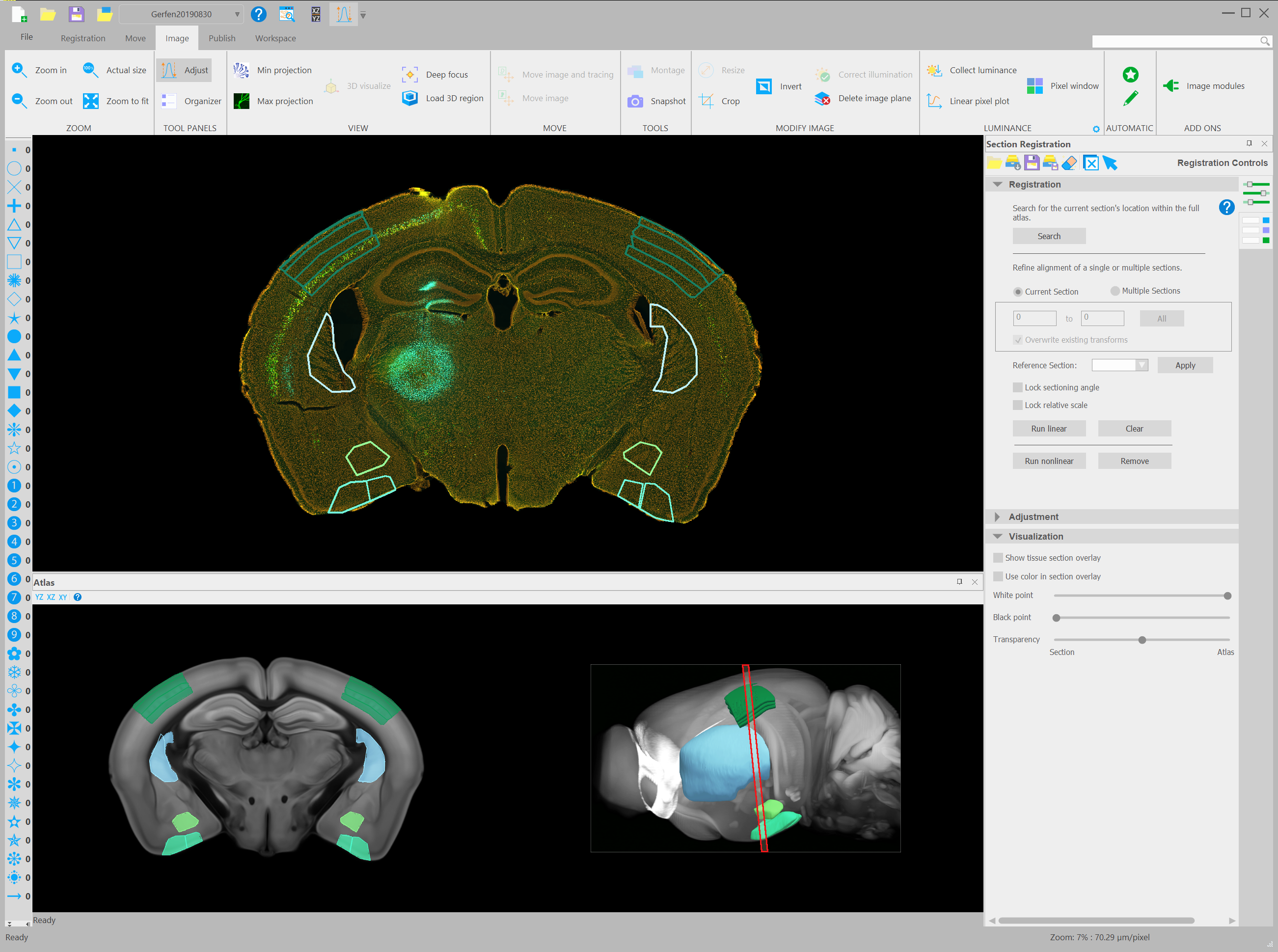
Coronal mouse brain section from the Laboratory of Systems Neuroscience of Dr. Charles Gerfen, NIMH Bethesda, Maryland
Meeting the demands of users is always a priority for MBF Bioscience, and working with customers like Dr. Charles Gerfen of the National Institute of Mental Health provided a major impetus for developing NeuroInfo into such a revolutionary product.
“The major advance,” said Dr. Gerfen, “is that we’re able to analyze projections from within and between different areas of the cerebral cortex to determine organizational principles of the cerebral cortex,” Along with Dr. Bryan M. Hooks of the University of Pittsburgh School of Medicine, Dr. Gerfen uses NeuroInfo to trace pyramidal neuron projections in Cre-driver mice (Hooks, et al 2018).
As outlined in a study published in Nature Communications, the research team first used MBF Bioscience’s BrainMaker functionality of NeuroInfo to reconstruct four Cre-recombinase driver mouse brains with sections imaged with Neurolucida. They then registered the reconstructed brains into the Allen Mouse Brain Atlas and then collectively visualized experimental data overlain with the atlas to determine exactly how the four populations of cortical pyramidal neurons they were tracking fit within the greater structure of the brain. “Essentially every pixel or image in our original images could be assigned to one of the 2500 brain structures in the Allen Atlas, said Dr. Gerfen.
Initially developed as a tool to help scientists automatically identify where their experimental tissue fits within specific regions of the mouse brain, NeuroInfo has evolved to encompass even more powerful methods for neuroanatomical analysis.
According to MBF Bioscience Senior Project Manager Dr. Nathan O’Connor, “scientists are using NeuroInfo most heavily to get their measurements mapped into a common coordinate system for objectively comparing and compiling results across animals, experiments, and laboratories. Rather than just a simple GPS — ‘where am I?’ tool, NeuroInfo has turned into a more powerful tool that allows automated mapping of brain-wide measures of behavior and pathology into a universal space.”
By automating the process of orientation, delineation, and quantification, the software cuts out intensive manual labor involved with traditional atlas use. It also provides a way for research teams to conduct large studies with less personnel, all while increasing repeatability and accuracy.
The key to the software’s success is a sophisticated algorithm that integrates with the Allen Mouse Brain Atlas, an extensive database of mouse brain anatomies delineated in a standardized average mouse brain [ref – https://mouse.brain-map.org/static/atlas]. Thanks to new developments within NeuroInfo, researchers can compare neuroanatomical data across mice, clearly viewing their areas of interest within the atlas and experimental data to observe how their specific data fits into the whole.
As explained in the paper, “Automatic navigation system for the mouse brain” (Tappan, et al 2019), NeuroInfo works much like a GPS, automatically navigating an investigator through the microscopic anatomy of histologic mouse brain sections. Once a slide is uploaded, the system registers and analyzes the data, then creates a digital overlay that clearly delineates the various brain regions contained within the section. As more and more labs integrate NeuroInfo into their research, the database grows, and researchers have more opportunities to enrich their studies.
After carrying out extensive validation studies, the research and development team, led by MBF Bioscience Scientific Director Dr. Susan Tappan, MBF Bioscience Imaging Scientist Dr. Brian Eastwood, and MBF Bioscience Senior Product Manager Dr. Nathan O’Connor, determined that the new technology identifies corresponding regions with remarkable accuracy. It also eliminates much of the manual labor that was previously required to identify the various brain regions in experimental sections.
“This novel automatic navigation system may fundamentally change the way researchers investigate sections of mouse brains under a light microscope, similar to the way surgical navigation systems have influenced neurosurgery over the past three decades (Mezger, Jendrewski and Bartels, 2013). Specifically, the approach presented here enhances the validity, reliability and reproducibility of studies that depend on the exact identification of distinct regions in the mouse brain, enables navigation through the complex anatomy of the mouse brain without requiring prior in-depth training in neuroanatomy, offers investigators the potential to analyze many more brain regions than is currently possible, and provides investigators with the means to directly correlate their experimental specimens with a number of reference tools such as GENSAT or the Allen Mouse Brain Connectivity Atlas (Oh et al., 2014),” Tappan, et al. 2019.
Citations:
Hooks BM, Papale AE, Paletzki RF, Feroze MW, Eastwood BS, Couey JJ, Winnubst J, Chandrashekar J, Gerfen CR (2018). “Topographic precision in sensory and motor corticostriatal projections varies across cell type and cortical area.” Nature Communications; doi: 10.1038/s41467-018-05780-7. https://www.nature.com/articles/s41467-018-05780-7
Tappan S, Eastwood BS, O’Connor N, Wang Q, Ng L, Feng D, Hooks B, Gerfen CR, Hof PR, Schmitz S, Glaser JR (2019). “Automatic navigation system for the mouse brain.” Journal of Comparative Neurology; 527(13):2200-2211. doi: 10.1002/cne.24635
https://www.ncbi.nlm.nih.gov/pubmed/30635922
Eastwood BS, Hooks BM, Paletzki RF, O’Connor NJ, Glaser JR, Gerfen CR (2018). “Whole Mouse Brain Reconstruction and Registration to a Reference Atlas with Standard Histochemical Processing of Coronal Sections.” Journal of Comparative Neurology; DOI: 10.1002/cne.24602
https://onlinelibrary.wiley.com/doi/abs/10.1002/cne.24602


